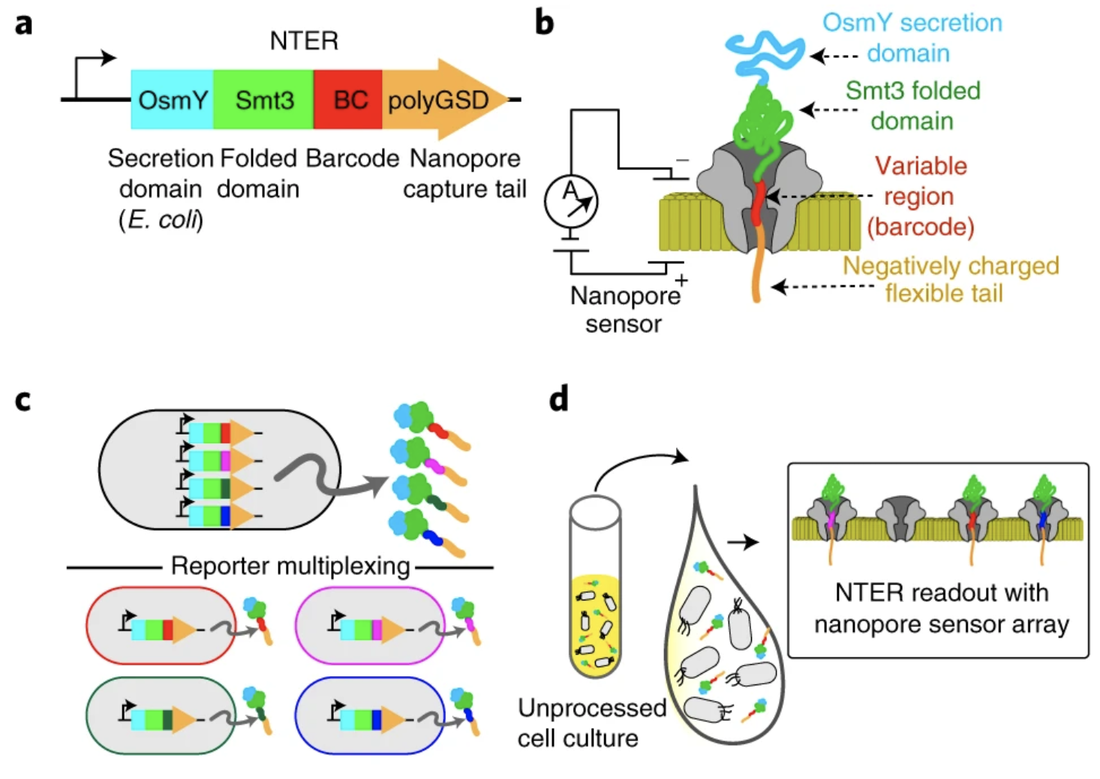
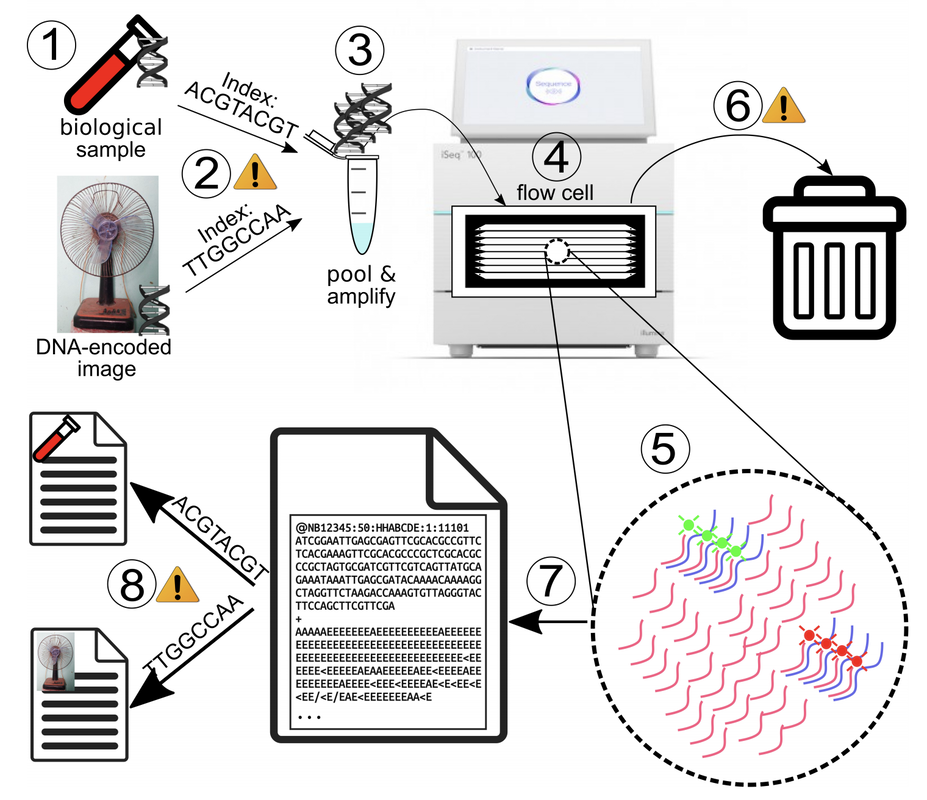
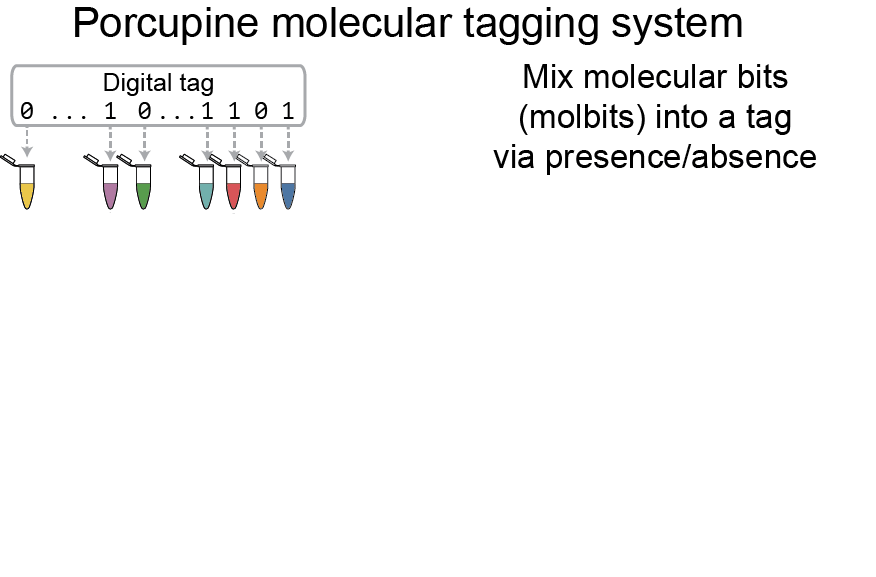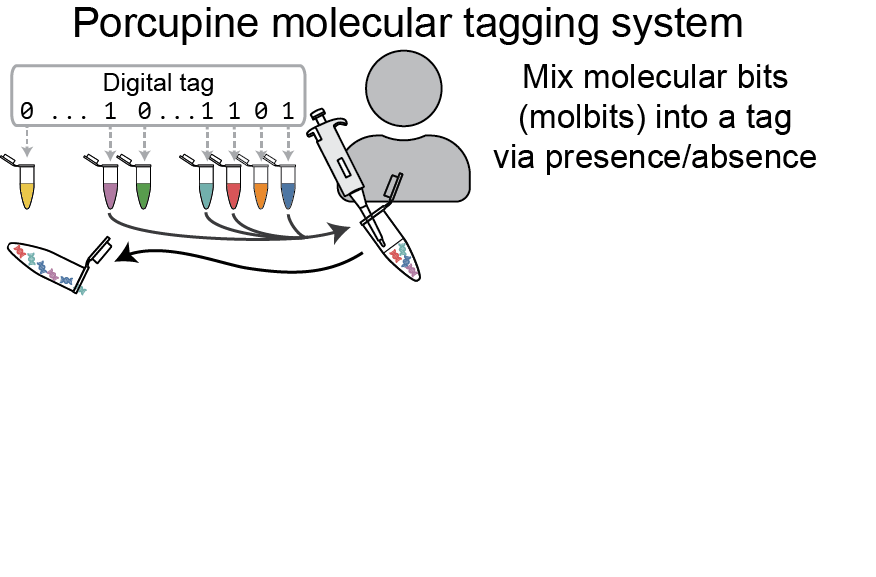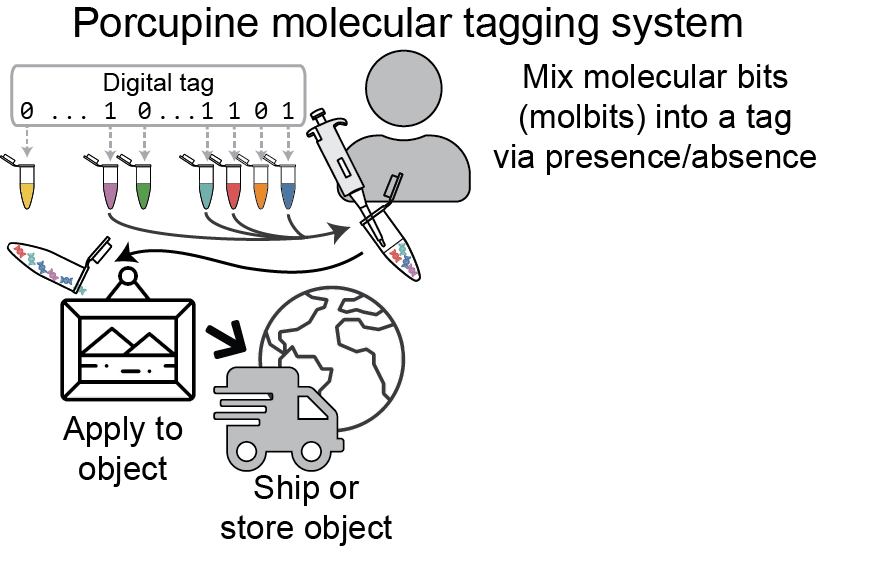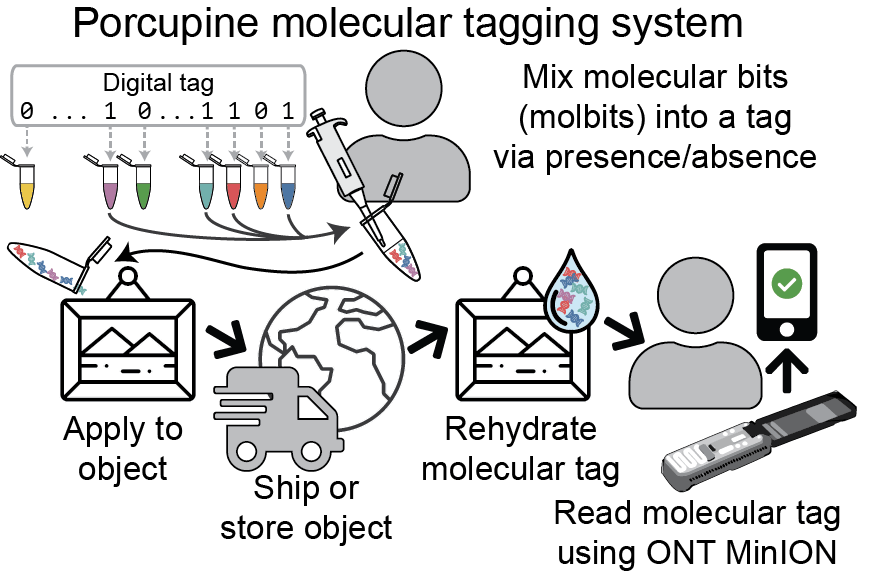
PREPRINTS & PUBLICATIONS
|
Vázquez Torres S, Leung PYJ, ..., Nivala J, Stewart L, Watson JL, Rogers JM, Baker D. De novo design of high-affinity binders of bioactive helical peptides. Nature. 626: 435–442. 2024.
Stephenson A, Lastra L, Nguyen B, Chen YJ, Nivala J, Ceze L, Strauss K. Physical Laboratory Automation in Synthetic Biology. ACS Synthetic Biology. 12 (11), 3156-3169. 2024. Motone K, Kontogiorgos-Heintz D, Wee J, Kurihara K, Yang S, Roote G, Fang Y, Cardozo N, Nivala J. Multi-pass, single-molecule nanopore reading of long protein strands with single-amino acid sensitivity. bioRxiv. 2023. Kuzdraliński A, Miśkiewicz M, Szczerba H, Mazurczyk W, Nivala J, Księżopolski B. Unlocking the potential of DNA-based tagging: current market solutions and expanding horizons. Nat. Comms. 14:6052. 2023. Stephenson A, Nivala J. Barcoding biomarkers with nanopore sequencing. Nat. Nanotechnol. 2023. Motone K, Nivala J. Not if but when nanopore protein sequencing meets single-cell proteomics. Nat. Methods. 20:336-338. 2023. Ney P, Bhattacharya A, Ceze L, Koscher K, Kohno T, Nivala J. Cybersecurity Across the DNA-Digital Boundary: DNA Samples to Genomic Data. Cyberbiosecurity. Book chapter. 95-114. 2023. Florian Praetorius*, Philip JY Leung*, Maxx H Tessmer, Adam Broerman, Cullen Demakis, Acacia F Dishman, Arvind Pillai, Abbas Idris, David Juergens, Justas Dauparas, Xinting Li, Paul M Levine, Mila Lamb, Ryanne K Ballard, Stacey R Gerben, Hannah Nguyen, Alex Kang, Banumathi Sankaran, Asim K Bera, Brian F Volkman, Jeff Nivala, Stefan Stoll, David Baker. Design of stimulus-responsive two-state hinge proteins. Science. 381:6659. 2023. Lakshmanam S, Leung PJY, Nivala J. Learning light-weight structure-aware embeddings for protein sequences. International Conference on Learning Representations (ICLR), Tiny Papers. 2023. Susana Vázquez Torres*, Philip JY Leung*, Isaac D Lutz, Preetham Venkatesh, Joseph L Watson, Fabian Hink, Huu-Hien Huynh, Andy Hsien-Wei Yeh, David Juergens, Nathaniel R Bennett, Andrew N Hoofnagle, Eric Huang, Michael J MacCoss, Marc Expòsit, Gyu Rie Lee, Paul M Levine, Xinting Li, Mila Lamb, Elif Nihal Korkmaz, Jeff Nivala, Lance Stewart, Joseph M Rogers, David Baker. De novo design of high-affinity protein binders to bioactive helical peptides. bioRxiv. 2022. Zhang K*, Chen YJ*, Delaney Wilde, Doroschak K, Strauss K, Ceze L, Seelig G, Nivala J. A nanopore interface for higher bandwidth DNA computing. Nat. Comms. 13:4904. 2022. Bhattarai-Kline S, Lear SK, Fishman CB, Lopez SC, Lockshin ER, Schubert MG, Nivala J, Church GM, Shipman SL. Recording gene expression order in DNA by CRISPR addition of retron barcodes. Nature. 2022. Ney P, Bhattacharya A, Ward D, Ceze LH, Kohno T, Nivala J. Doctoring Direct-to-Consumer Genetic Tests with DNA Spike-Ins. bioRxiv. 2022. Meiser LC, Nguyen BH, Chen YJ, Nivala J, Strauss K, Ceze L, Grass RN. Synthetic DNA applications in information technology. Nat. Comms. 13: 352. 2022. Zhang K*, Chen YJ*, Doroschak K, Strauss K, Ceze L, Seelig G, Nivala J. A nanopore interface for high bandwidth DNA computing. bioRxiv. 2021. Motone K, Cardozo N, Nivala J. Herding cats: label-based approaches in protein translocation though nanopore sensors for single-molecule protein sequence analysis. iScience. 2021. Cardozo N*, Zhang K*, Doroschak K*, Nguyen A, Siddiqui Z, Bogard N, Strauss K, Ceze L, Nivala J. Multiplexed direct detection of barcoded protein reporters on a nanopore array. Nat. Biotechnol. 2021. News and highlight coverage:
Ney P, Organick L, Nivala J, Ceze L, Kohno T. DNA sequencing flow cells and the security of the molecular digital interface. Privacy Enhancing Securities Symposium. 2021. Doroschak K, Zhang K, Queen M, Mandyam A, Strauss K, Ceze L, Nivala J. Rapid and robust assembly and decoding of molecular tags with DNA-based nanopore signatures. Nat. Comms. 11, Article number: 5454. 2020. preprint: https://www.biorxiv.org/content/10.1101/2020.03.06.981514v1 Nivala J. Perspective: Follow the barcoded microbes. Science. 2020. Cardozo N*, Zhang K*, Doroschak K, Nguyen A, Siddiqui Z, Strauss K, Ceze L, Nivala J. Multiplexed direct detection of barcoded protein reporters on a nanopore array. 2019. https://www.biorxiv.org/content/10.1101/837542v1 Ceze L, Nivala J, Strauss K. Molecular digital data storage using DNA. Nat. Rev. Genet. 2019. (Review Article) Nivala J, Shipman SL, Church GM. Spontaneous CRISPR-loci generation in-vivo by non-canonical spacer integration. Nat. Microbiol. (3):310-318. 2018.
Shipman SL, Nivala J, Macklis JD, Church GM. CRISPR-Cas encoding of a digital movie into the genomes of a population of living cells. Nature. 547. 345-349. PMID: 28700573. 2017.
Pallo M*, Stranges PB*, Kalachikov S, Nivala J, Dorwart M, Trans A, Kumar S, Porel M, Chien M, Tao C, Morozova I, Li Z, Shi S, Aberra A, Arnold C, Yang A, Aguirre A, Harada ET, Korenblum D, Pollard J, Bhat A, Gremyachinskiy D, Bibillo A, Chen R, Davis R, Russo JJ, Fuller CW, Roever S, Ju J, and Church GM. Design and characterization of a nanopore-coupled polymerase for single molecule DNA sequencing by synthesis on an integrated electronic array. PNAS. 113(19):5233-8. PMID: 27091962. 2016. Shipman SL*, Nivala J*, Macklis JD, Church GM. Molecular recordings by directed CRISPR spacer acquisition. Science. 353(6298). PMID 27284167. 2016. *equal contribution
Nivala J, Mulroney L, Li Gf, Schreiber J, Akeson M. Discrimination among protein variants using an unfoldase-coupled nanopore. ACS Nano. 23;8(12):12365-75. PMID: 25402970. 2014. Nivala J, Marks DB, Akeson M. Unfoldase-mediated protein translocation through an α-hemolysin nanopore. Nat. Biotechnol. (3):247-50. PMID: 23376966. 2013.
Wu SJ, Eiben CB, Carra JH, Huang I, Zong D, Liu P, Wu CT, Nivala J, Dunbar J, Huber T, Senft J, Schokman R, Smith MD, Mills JH, Friedlander AM, Baker D, Siegel JB. Improvement of a potential anthrax therapeutic by computational protein design. J Biol Chem. 286(37):32586-92. PMID: 21768086. 2011. Actis P, Rogers A, Nivala J, Vilozny B, Seger RA, Jejelowo O, Pourmand N. Reversible thrombin detection by aptamer functionalized STING sensors. Biosens Bioelectron. 26(11):4503-7. PMID: 21636261. 2011. |
Book chapters & Additional Pubs
Nivala J*, Mulroney L, Luan Q, Abu-Shumays R, Akeson M. Unfolding and translocation of proteins through an alpha-hemolysin nanopore by ClpXP. Methods in Molecular Biology. 2021;2186:145-155. PMID: 32918735. 2020. *corresponding author
Shipman SL and Nivala J. Generating molecular records in the genome. Cell Systems (Principles of Systems Biology No. 7) 1, 307. 2016.
Shipman SL and Nivala J. Generating molecular records in the genome. Cell Systems (Principles of Systems Biology No. 7) 1, 307. 2016.